How I start a bioinformatics project
I started analyzing sequencing data just over one year ago. In the beginning it was so exciting to finally receive the first reads, do some quality checks and start further analysis, that I didn´t think about reproducibility. I ended up with multiple distinct versions of mapped reads, adapter/quality trimmed reads from scripts using different parameters. It was bad…
So I had to force myself to rethink my workflow, which I will show you (in a simplifed way) in the following.
My personal workflow
Of course I`m not the first one who had this problem and if you are new to bioinformatics and sequencing you will probably encounter similar difficulties when starting your analysis. There are probably many ways you can achieve a certain level of organization without the need for any advanced technique, like version control. Sooner or later you´ll get there automatically. Trust me.
Here, I will show you how to organize yourself using RStudio.
First, open a new window and create a new project.
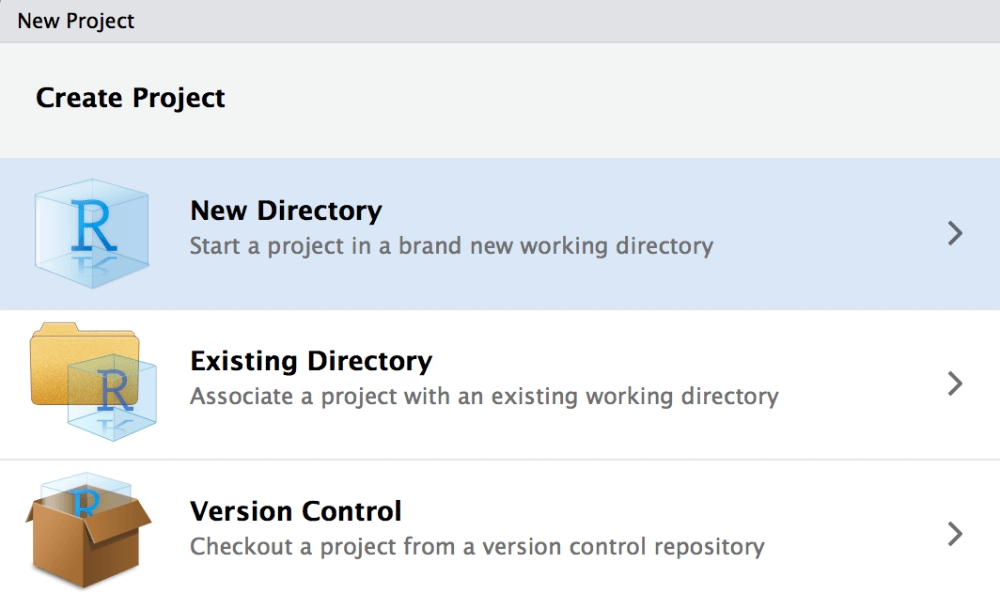
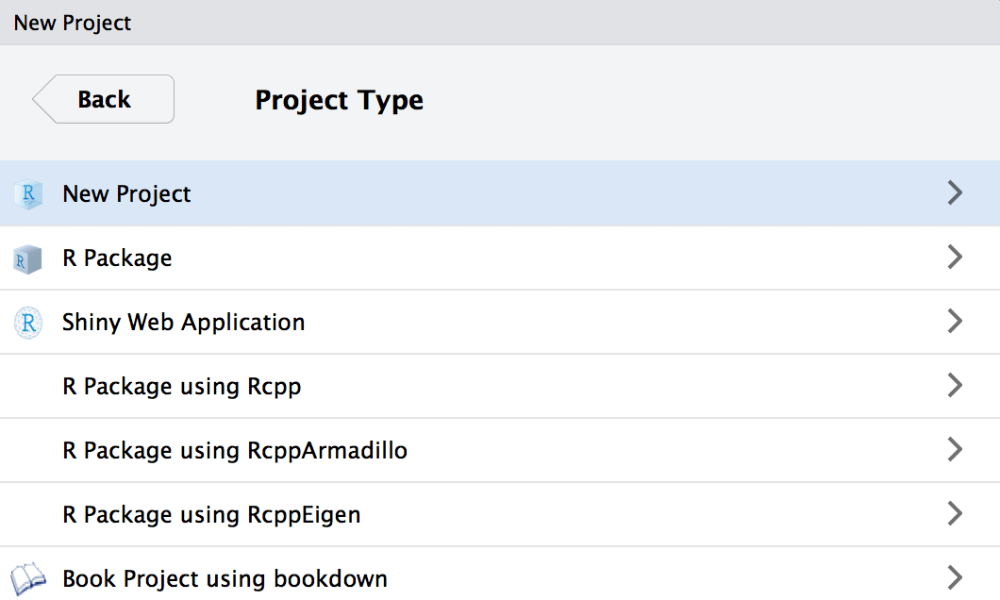
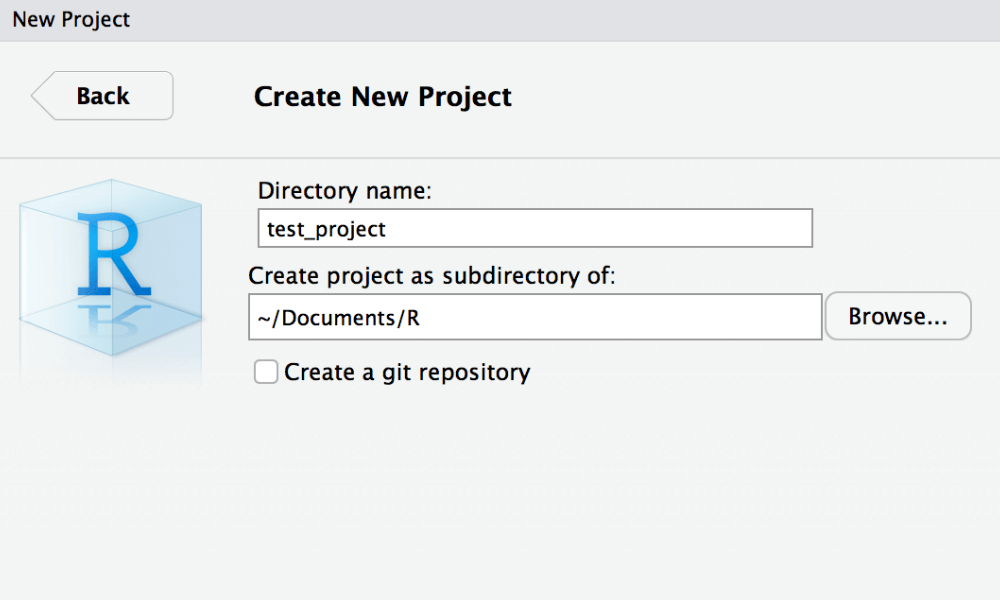
Within the new RStudio window switch to the embedded terminal (new feature since version 1.1). If you are not sure about the version you are actually using you can check this with typing RStudio.Version() in the RStudio console.
Since you are already in your new project folder, we can start be making 4 folders for your analysis.
# in RStudio terminal
project_folder=${PWD}
mkdir -p ${project_folder}/{data,scripts,results,doc}
Once you have done that, simply save your input sequencing data in data and so on. For example you can put genome annotation data you use for your analysis in data/genome_data.
Since you´re already in RStudio it is quite easy to open an RMarkdown document to keep track of your code and also thoughts about the different steps (doc). The remaining two folder scripts and results are self-explanatory, but important for your organization so that input files do not get mixed up with annotation or any other results files of a following step.
Using Rmarkdown and Rproject package
Actually there is an Rpackage called ProjectTemplate that is perfectly suited to start any data analysis project. In case you do not want to read full documentation here is how you get started:
# in R
install.packages("ProjectTemplate")
library("ProjectTemplate")
create.project("../test_project", merge.strategy = "allow.non.conflict")
setwd("~/test_project")
library('ProjectTemplate')
load.project()
Key message
I keep it simple: Make folders or you get lost…
In case you want to get more information you can start by reading a paper published in PLOS computationsl biology. Feel free to comment on my workflow.

Leave a Comment